Alan Wardroper – MRes Bioinformatics
University of York
Autumn 2003 Project Report
Using
Minimal Genomes to Establish the Importance of Uncharacterised Genes in E.
coli
Supervisors:
Prof. Angela Douglas & Dr. Gavin Thomas
2,155
words in text, References, 4 Tables, 6 Figures, 4 Appendices
Background and Aims
Despite the long history of use of Escherichia coli
as a biological model system, the sequencing of its genome (Blattner et al.,
1997) revealed a number of surprises – not least of which was the discovery of
more than 2000 open reading frames (orfs)
encoding potential products of unknown function. These as yet uncharacterised
genes, provisionally given y-names based on their map position (Rudd, 1998),
account for more than 50% of the E. coli genome, which naturally raises
questions regarding their functions.
One approach to address the relative importance of these ORFs is to search for homologues in closely related species with reduced genomes (Table 1).
Table 1.
Comparison of Bacterial Genome Lengths.
| Organism |
Genome
Length |
Accession |
| Escherichia coli K12 |
4639221
bp |
NC_000913 |
| Salmonella typhimurium LT2 |
4,857,432
bp |
NC_003197
|
| Yersinia pestis strain CO92 |
4653728
bp |
NC_003143
|
| Klebsiella pneumoniae |
4893316
bp |
NC_002941 |
| Buchnera
aphidicola str. APS (Acyrthosiphon pisum) |
640681
bp |
NC_002528
|
| Buchnera
aphidicola str. Sg (Schizaphis graminum) |
641454
bp |
NC_004061
|
| Buchnera
aphidicola str. Bp (Baizongia pistaciae) |
615980
bp |
NC_004545
|
| Wigglesworthia glossinidia (endosymbiont of Glossina
brevipalpis) |
697724
bp |
NC_004344
|
| Candidatus Blochmannia floridanus |
705557
bp |
NC_005061
|
| Mycoplasma genitalium |
580,074
bp |
NC_000908 |
Genome reduction
Mitochondria and chloroplasts are thought to have evolved
from free-living microorganisms through endosymbiotic events including genome
reduction. Many of the endosymbiont genes would have been redundant once in
the environment of a host cell and were since lost through deletion or inactivation
due to relaxation of selective pressure. Other genes have been transferred
from the endosymbiont to the host genome, the litmus test of true organelle
status (Douglas & Raven, 2003). As shown in Fig. 1, the free-living species
E. coli and Y. pestis contain significantly greater numbers
of uncharacterised genes than Buchnera (B.Ap) or M. genitalium,
both species with greatly reduced genomes (p<0.001).
Fig. 1.
Functional Classification of Bacterial Genes
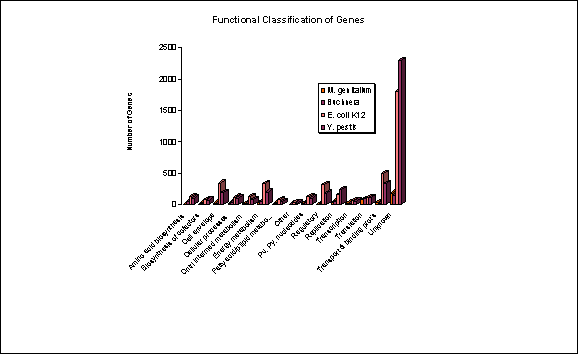
Comparison of functional gene classifications among bacterial
species. As shown in the chart, the free-living species E. coli and
Y. pestis contain significantly greater numbers of uncharacterised
genes than Buchnera (B.Ap) or M. genitalium, both species with
greatly reduced genomes (p<0.001).
(figures/Fig1FuncClass.xls)
Buchnera
Buchnera are bacterial species
closely related to the Enterobacteriaceae, diverging from a common
ancestor with E. coli between 200 and 150 million years ago (MYA), since
which time they have become obligate mutualistic intracellular symbionts of a
number of aphid species. Despite their close relation to E. coli, Buchnera
have undergone marked genome reduction, with genome lengths of only about
13% of those of their free-living relatives (Table 1). Interestingly, neither Buchnera
not the tsetse fly endosymbiont Wigglesworthia glossinidia appear to possess significant
numbers of unique genes not present in free-living enterobacteria (only 4 and
3, respectively). Genes retained throughout evolution in organisms with such
drastically reduced genomes are likely to be crucial for life. Therefore, this
study was performed to determine which of the uncharacterised genes in E.
coli are most important based on their retention in Buchnera and
other closely related endosymbionts with reduced genomes.
Methods
To
determine the importance of the uncharacterised ORFs in E. coli based on
their retention in organisms that have undergone genome reduction, the subsets
of these ORFs retained in three species of Buchnera – B. aphidicola str. Acyrthosiphon pisum (B.Ap), B. aphidicola str. Baizongia pistaciae (B.Bp), and B. aphidicola str. Schizaphis graminum (B.Sg) – were
first compared. Then, the overlaps with the subsets retained in other species
with reduced genomes (the carpenter ant endosymbiont Candidatus Blochmannia
floridanus, and Wigglesworthia glossinidia, an endocellular obligate
endosymbiont of the tsetse fly Glossina brevipalpis) were determined to further refine the search for
crucial genes among these candidates.
Gene tables were obtained from NCBI < http://www.ncbi.nlm.nih.gov/> and
analysed using the Unix regular expression pattern-matching tool grep to search for genes annotated as hypothetical,
putative, possible, unknown etc. These lines were piped to result files,
which were then opened for analysis in MS Excel 2000 and Sun OpenOffice Calc.
A local BLAST v 2.2.6 installation <http://www.ncbi.nlm.nih.gov/BLAST/> was
used to simultaneously compare multiple sequences among various combinations of
species.
The Artemis Comparison Tool (ACT) DNA
sequence comparison analysis viewer Release 2 <http://www.sanger.ac.uk/Software/ACT/> was
used to examine synteny between each of the Buchnera species and E.
coli using the BLAST output.
Commands used:
Grep to find hypothetical/characterised annotations:
$>grep
–[e | v] hypot input_filename > output_filename ; grep –e
putat input_filename >> output_filename; grep –e possible input_filename
>> output_filename
BLAST database preparation:
$>formatdb
–i <input_filename.fas> -p F
Produce BLAST tables for ACT:
$>blastall
–p blastn –d <database_name> -i <query_filename> -e
[0.1 | 0.9] –m 8 –J T –p F –o <output_filename>
Table 2 shows a comparison of the core uncharacterised Buchnera genes with the Yersinai pestis genome
Table 2.
Results of wublastp of
Core Uncharacterised Buchnera genes vs. Yersinia pestis genome
| Query |
Buchnera |
Y.p name |
Score: |
(P/N):
|
N:
|
| Glob |
BU246 |
YPO1079 |
573 |
1.50E-57 |
1 |
| RnfA |
BU113 |
YPO2246 |
587 |
4.90E-59 |
1 |
| SohB |
BU283 |
YPO2216 |
788 |
9.90E-84 |
2 |
| YabC |
BU224 |
YPO0547 |
782 |
1.10E-79 |
1 |
| YabI |
BU139 |
YPO0519 |
622 |
9.50E-63 |
1 |
| YajC |
BU134 |
yajC |
319 |
1.20E-30 |
1 |
| YajR |
BU466 |
yajR |
588 |
7.20E-69 |
2 |
| YbaB |
BU482 |
YPO3121 |
353 |
3.10E-34 |
1 |
| YbeD |
BU488 |
YPO2600 |
251 |
2.00E-23 |
1 |
| YbeY |
BU442 |
YPO2618 |
302 |
7.70E-29 |
1 |
| YbgI |
BU301 |
YPO2697 |
704 |
2.00E-71 |
1 |
| YbhE |
BU293 |
YPO1149 |
518 |
1.00E-51 |
1 |
| YccK |
BU467 |
YPO1447 |
319 |
1.20E-30 |
1 |
| YceA |
BU365 |
YPO2451 |
939 |
2.40E-96 |
1 |
| YcfF |
BU357 |
YPO1611 |
288 |
2.40E-27 |
1 |
| YcfH |
BU355 |
YPO1607 |
644 |
4.40E-65 |
1 |
| YchE |
BU267 |
YPO2181 |
598 |
3.30E-60 |
1 |
| YchF |
BU191 |
ychF |
1025 |
1.90E-105 |
1 |
| YciA |
BU274 |
YPO2195 |
434 |
8.00E-43 |
1 |
| YciC |
BU276 |
YPO2199 |
218 |
6.20E-20 |
1 |
| YdgO |
BU116 |
YPO2242 |
599 |
2.60E-60 |
1 |
| YdgQ |
BU118 |
YPO2240 |
490 |
9.30E-49 |
1 |
| YdiC |
BU122 |
YPO2404 |
152 |
6.10E-13 |
1 |
| YeaZ |
BU324 |
YPO2072 |
275 |
5.60E-26 |
1 |
| YfgB |
BU286 |
YPO2882 |
1005 |
2.50E-103 |
1 |
| YfgM |
BU608 |
yfgM |
132 |
8.00E-11 |
1 |
| YfhC |
BU255 |
YPO2923 |
352 |
3.90E-34 |
1 |
| YfjF |
BU253 |
YPO1103 |
221 |
3.00E-20 |
1 |
| YgfZ |
BU435 |
YPO0898 |
564 |
1.30E-56 |
1 |
| YggB |
BU452 |
YPO0919 |
796 |
3.50E-81 |
1 |
| YggH |
BU551 |
YPO0951 |
506 |
1.90E-50 |
1 |
| YggS |
BU549 |
YPO0941 |
368 |
7.90E-36 |
1 |
| YggW |
BU550 |
YPO0946 |
949 |
2.10E-97 |
1 |
| YggX |
BU553 |
YPO0953 |
256 |
5.80E-24 |
1 |
| YheL |
BU530 |
YPO0199 |
202 |
3.10E-18 |
1 |
| YheM |
BU531 |
YPO0198 |
248 |
4.10E-23 |
1 |
| YheN |
BU532 |
YPO0197 |
317 |
2.00E-30 |
1 |
| YhfC |
BU535 |
YPO0163 |
1060 |
3.70E-109 |
1 |
| YhgI |
BU544 |
YPO0127 |
510 |
7.00E-51 |
1 |
| YhgN |
BU449 |
yhgN |
501 |
6.30E-50 |
1 |
| YibN |
BU052 |
YPO0065 |
247 |
5.20E-23 |
1 |
| YigL |
BU028 |
YPO3829 |
411 |
2.20E-40 |
1 |
| YleA |
BU441 |
YPO2620 |
1435 |
6.70E-149 |
1 |
| YnfM |
BU588 |
YPO2266 |
849 |
8.40E-87 |
1 |
| YoaE |
BU323 |
yoaE |
1184 |
2.70E-122 |
1 |
| YqgF |
BU548 |
YPO0937 |
360 |
5.50E-35 |
1 |
| YraL |
BU091 |
YPO3547 |
773 |
9.50E-79 |
1 |
| YrbA |
BU385 |
YPO3570 |
186 |
1.50E-16 |
1 |
| YrdC |
BU494 |
YPO0245a |
415 |
8.20E-41 |
1 |
No homologue of yba2 in Y. pestis
Analysis of the Candidate Genes
SignalP
<http://www.cbs.dtu.dk/services/SignalP/> was used to predict
the presence of signal peptides, as a preliminary screen to search for possible
secretory/periplasmic proteins.
Predictions were made regarding the subcellular
localisations of the predicted protein products using PSORT
<http://psort.nibb.ac.jp/>
HMMTop <http://www.enzim.hu/hmmtop/> was
used to predict topology of the putative gene products.
PROSITE <http://us.expasy.org/prosite/> was
used to examine the predicted protein sequences encoded by the uncharacterised
ORFs for common structural motifs to gain insight into their possible
functions.
InterPro < http://www.ebi.ac.uk/interpro/>
was used to scan each sequence for known protein family signatures, domains and
functional sites.
The results of these analyses are summarised in Table 3.
Table 3.
Core Uncharacterised Genes Common to all Three Buchnera Species: Motif, Localisation,
and Topology Predictions
| Gene |
Signal P prediction |
Motifs
(PROSITE) |
Localisation (PSORT
prediction) |
HMMTop |
|
| N-Ter |
T/M Helix |
||||
| gloB |
NS |
Prokar_Lipoprotein |
bacterial outer membrane 0.724: Affirmative ) |
OUT |
0 |
| rnfA |
NS |
Prokar_Lipoprotein |
bacterial inner membrane (0.157: Affirmative ) |
OUT |
6 |
| rnfD/ydgO |
NS |
|
bacterial inner membrane (0.444: Affirmative ) |
OUT |
9 |
| rnfE/ydgQ |
SP
|
|
bacterial inner membrane (0.351: Affirmative ) |
OUT |
6 |
| sohB |
NS |
Fabp |
bacterial outer membrane (0.790: Affirmative ) |
IN |
3 |
| yabC |
NS |
|
bacterial cytoplasm (0.256: Affirmative ) |
OUT |
0 |
| yabI |
NS |
Prokar_Lipoprotein |
bacterial cytoplasm (0.392: Affirmative ) |
IN |
5 |
| yajC |
NS |
|
bacterial cytoplasm (0.215: Affirmative ) |
OUT |
1 |
| yajR |
NS |
|
bacterial inner membrane (0.588: Affirmative ) |
IN |
12 |
| yba2 |
NS |
|
bacterial cytoplasm (0.261: Affirmative ) |
OUT |
0 |
| ybaB |
NS |
|
bacterial cytoplasm (0.352: Affirmative ) |
OUT |
0 |
| ybeD |
NS |
|
bacterial cytoplasm (0.167: Affirmative ) |
IN |
1 |
| ybeY |
NS |
Crystallin_Betagamma, Upf0004 |
bacterial cytoplasm (0.038: Affirmative ) |
OUT |
0 |
| ybgI |
NS |
|
bacterial cytoplasm (0.381: Affirmative ) |
OUT |
0 |
| ybhE |
NS |
|
bacterial cytoplasm (0.450: Affirmative ) |
OUT |
0 |
| yccK |
NS |
|
bacterial cytoplasm (0.443: Affirmative ) |
OUT |
0 |
| yceA |
NS |
|
bacterial cytoplasm (0.438: Affirmative ) |
OUT |
0 |
| ycfF |
NS |
|
bacterial cytoplasm (0.298: Affirmative ) |
OUT |
0 |
| ycfH |
NS |
Upf0006_1, Upf0006_2 |
bacterial inner membrane (0.601: Affirmative ) |
OUT |
0 |
| ychE |
NS |
|
bacterial inner membrane (0.497: Affirmative ) |
OUT |
6 |
| ychF |
NS |
Atp_Gtp_A Gatase_Type_Ii |
bacterial cytoplasm (0.062: Affirmative ) |
OUT |
0 |
| yciA |
NS |
|
bacterial cytoplasm (0.054: Affirmative ) |
IN |
3 |
| yciC |
NS |
|
bacterial inner membrane (0.537: Affirmative ) |
IN |
6 |
| ydiC |
NS |
|
bacterial cytoplasm (0.336: Affirmative ) |
OUT |
2 |
| yeaZ |
NS |
|
bacterial inner membrane (0.191: Affirmative ) |
OUT |
0 |
| yfgB |
NS |
|
bacterial cytoplasm (0.242: Affirmative ) |
OUT |
0 |
| yfgM |
SP |
|
bacterial inner membrane (0.631: Affirmative ) |
OUT |
2 |
| yfhC |
NS |
|
|
OUT |
2 |
| yfjF |
NS |
|
bacterial inner membrane (0.143: Affirmative ) |
OUT |
0 |
| ygfZ |
NS |
Prokar_Lipoprotein |
bacterial inner membrane (0.601: Affirmative ) |
OUT |
0 |
| yggB |
NS |
|
bacterial inner membrane (0.359: Affirmative ) |
OUT |
4 |
| yggH |
NS |
|
bacterial cytoplasm (0.193: Affirmative ) |
OUT |
0 |
| yggS |
NS |
Upf0001 |
bacterial cytoplasm (0.397: Affirmative ) |
OUT |
0 |
| yggW |
NS |
|
bacterial cytoplasm (0.217: Affirmative ) |
OUT |
0 |
| yggX |
NS |
|
bacterial cytoplasm (0.126: Affirmative ) |
OUT |
0 |
| yheL |
NS |
|
bacterial inner membrane (0.073: Affirmative ) |
OUT |
0 |
| yheM |
NS |
|
bacterial inner membrane (0.138: Affirmative ) |
IN |
1 |
| yheN |
NS |
|
|
OUT |
0 |
| yhfC |
SP |
|
bacterial cytoplasm (0.394: Affirmative ) |
IN |
12 |
| yhgI |
NS |
Prokar_Lipoprotein |
bacterial cytoplasm (0.313: Affirmative ) |
OUT |
0 |
| yhgN |
NS |
|
bacterial inner membrane (0.525: Affirmative ) |
IN |
5 |
| yibN |
MA |
|
bacterial cytoplasm (0.179: Affirmative ) |
OUT |
2 |
| yigL |
NS |
Cof_1 |
bacterial cytoplasm (0.217: Affirmative ) |
OUT |
0 |
| yleA |
NS |
|
bacterial cytoplasm (0.038: Affirmative ) |
OUT |
0 |
| ynfM |
SP |
Prokar_Lipoprotein, Sugar_Transport_1 |
bacterial inner membrane (0.539: Affirmative ) |
IN |
12 |
| yoaE |
NS |
|
bacterial inner membrane (0.631: Affirmative ) |
OUT |
7 |
| yqgF |
NS |
|
bacterial inner membrane (0.109: Affirmative ) |
OUT |
0 |
| yraL |
NS |
|
bacterial cytoplasm (0.107: Affirmative ) |
OUT |
2 |
| yrbA |
NS |
|
bacterial inner membrane (0.499: Affirmative ) |
OUT |
0 |
| yrdC |
NS |
Sua5 |
bacterial cytoplasm (0.330: Affirmative ) |
OUT |
0 |
Shaded columns indicate core uncharacterised genes common
to all three Buchnera species, Wigglesworthia and C. Blochmannia.
NS, Non-secretory
protein predicted by SignalP; PP/Sc, Periplasmic/secreted protein; MA, membrane-anchored
protein (see adjacent poster by Louise Fairweather); SP, Signal peptide predicted by SignalP.
Statistical analyses
Differences in percentage of uncharacterised genes
between species/groups were analysed by χ2 test, and
differences in gene length between uncharacterised and annotated genes were
analysed by one-way ANOVA. Values of p<0.05 were taken to indicate
significance. Statistical analyses were performed using Microsoft Excel 2000
and SPSS 11.
Results
After normalisation by name, analysis revealed 97
uncharacterised genes in B.Ap, 95 in the closely related species B.Sg,
and 86 in the more distantly related sister species B.Bp. Of these
hypothetical genes, 50 were common to all three Buchnera species. In addition
to these 50, B.Ap shared 26 and 10 uncharacterised genes with B.Sg
and B.Bp, respectively, while B.Bp and B.Sg shared 10 with
each other but not B.Ap (Table 3, Fig. 2).
Fig. 2.
Distribution of Uncharacterised Genes Among Buchnera
Species.
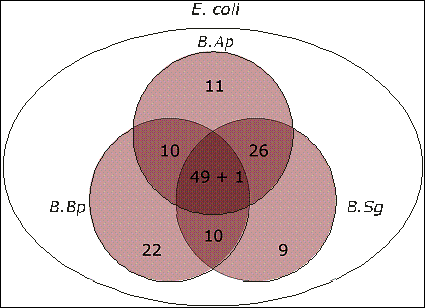
The three species B.Ap, B.Sg, and B.Bp
shared 49 of the uncharacterised ORFs from E. coli as well as 1 hypothetical
gene apparently unique to Buchnera.
(figures/Fig2venn1.cdr)
As shown in Fig. 2, the superset of uncharacterised
genes in Buchnera includes 138 genes,
all but one of which are present in E. coli. These 138 genes were presumably
all present in the common ancestor of the three Buchnera species, with subsets being lost through genome reduction
as a result of reduced selective pressure.
Of the 50 core genes shared by all three Buchnera species, 29 (58%) were also common
to Wigglesworthia glossinidia an obligate endosymbiont of the tsetse
fly Glossina brevipalpis, and 23 were common to Candidatus Blochmannia.
A subset of 20 of the 50 uncharacterised genes common to all three Buchnera
species were shared with both Candidatus Blochmannia floridanus, and
Wigglesworthia glossinidia (Figs. 2,3, Table 3).
Fig. 3.
Distribution of Uncharacterised Genes Among Endosymbionts.
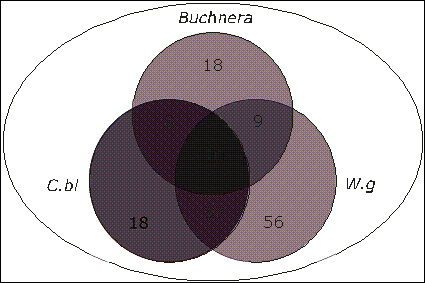
Of the 49 hypothetical genes common to all three Buchnera species, 20 were also found in
Candidatus Blochmannia floridanus and Wigglesworthia glossinidia.
(figures/Fig3venn2.cdr)
Synteny
The loci corresponding to the 20 core uncharacterised
E. coli ORFs in the 5 endosymbiont species were plotted as a function
of chromosome position. The results indicated that gene order and chromosomal
locations of the uncharacterised ORFs are well conserved among all three Buchnera species (Fig. 4), but highly diverged
between Buchnera and the other two endosymbiont species with similar
genome sizes. This is remarkable given that the overall degree of synteny,
as demonstrated by ACT analyses, was markedly high only between B.Ap and B.Sg, with a much lesser degree of conservation of gene order between
these two strains and B.Bp (Fig.
5). The core uncharacterised genes lie in the same order along the chromosomes
and are in similar locations in all three Buchnera species; all five
endosymbionts are plotted in Fig. 5 but the degree of agreement is so high
between B.Ap and B.Sg that they are essentially superposed over most of their length.
The degree of alignment between B.Bp and the other two species was
greatest nearer the origin of replication, and showed increasing divergence
with distance. The lines began to become well separated around 378,000, possibly
reflecting a positional bias along the genome for gene deletion/degradation
with stronger constraints around the origin.
Fig. 4.
Phylogenetic Relationships between Buchnera and Free-Living γ-Proteobacteria.
Figure adapted from Moran and Miura, Genome Biology
2, No 12, 2001
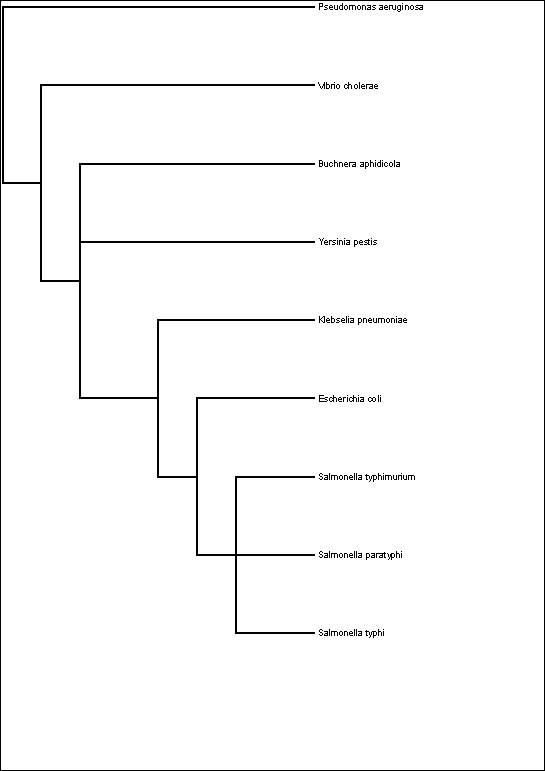
Phylogenetic tree showing the evolutionary relationships
between Buchnera and γ-Proteobacteria.
(figures/Fig4phyloTree.wmf derived from phyloTree.tre)
Taken together, these observations suggest that
extensive chromosomal rearrangements have occurred since the separation of the
lineages of Blochmannia, Wigglesworthia and Buchnera from their
common ancestor, but that the extent of such large-scale alterations in Buchnera
is small, consistent with the high
degree of chromosomal stability in endosymbionts reported previously (Tamas et
al., 2002; Silva et al., 2003). The trend line in Fig 5
also indicates that these hypothetical genes are dispersed relatively evenly
along the whole length of the Buchnera
genome. This is again remarkable as the results of ACT analysis shown in Fig. 4
suggested that the endosymbiotic evolution of Buchnera involved a greater degree of gene disintegration and loss
from the central regions of the common ancestor with E. coli.
The loci corresponding to the 20 core uncharacterised E.
coli ORFs in 5 endosymbiont species were plotted as a function of
chromosome position. The plots clearly show good conservation of both order and
position of these uncharacterised genes among the Buchnera species. The
lines for B.Ap and B.Sg are superposed over most of the
plot, but a good match was also found for the more distantly related B.Bp.
Fig. 5.
Synteny in Endosymbionts.
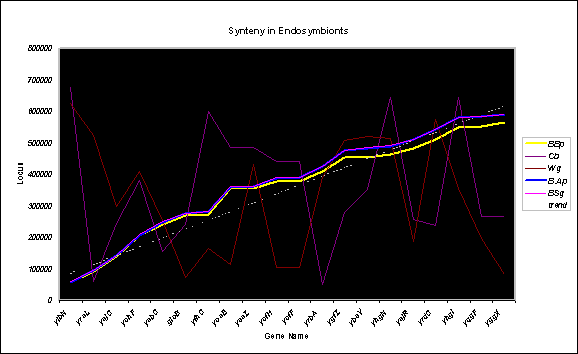
The loci corresponding to the 20 core uncharacterised
E. coli ORFs in 5 endosymbiont species were plotted as a function of
chromosome position. The plots clearly show good conservation of both order
and position of these uncharacterised genes among the Buchnera species.
The lines for B.Ap and B.Sg are superposed over most of the plot,
but a good match was also found for the more distantly related B.Bp. The linear trend line indicates essentially
homogeneous distribution along the chromosome. The other two endosymbionts
show markedly different gene orders and positions.
B.Ap, Buchnera
aphidicola str. Acyrthosiphon pisum; B.Bp, B. aphidicola str. Baizongia pistaciae;
B.Sg, B. aphidicola str. Schizaphis graminum; Cb, Candidatus Blochmannia floridanus;
Wg, Wigglesworthia glossinidia.
Comparison with Yersinia pestis
Based on the annotation, only 2 of the 50 core
hypothetical genes, gloB and sohB,
were also common to the free-living plague enterobacterium Yersinia pestis,
chosen due to its close phylogenetic relationship to both Buchnera and E.
coli (Fig. 4). The annotation for this bacterium is extremely sparse, with
almost 1000 hypothetical genes having no meaningful designation. Therefore,
translated tblastn BLAST searches were
performed against the Y. pestis strain CO92 complete genome sequence
<http://www.sanger.ac.uk/Projects/Y_pestis/>,
and significant hits were obtained for all but one of the 50 core hypothetical Buchnera genes. These observations
underlie the need for not only a greater degree of standardisation in
annotation but also for caution in interpretation of data based on potentially
misleading annotation.
The one gene that was not represented in the Yersinia
genome was the Buchnera-specific
gene yba2, which also not present in E. coli, Blochmannia,
or Wigglesworthia .
Gene Characteristics
The uncharacterised genes in B.Ap are on the
whole short, with an average gene length of 684.76 bp.This is in contrast to
the average overall gene length of 937.55bp for the genome as a whole, a figure
very close to the accepted average bacterial protein-coding
gene length of 1 kb. In contrast, the value for the hypothetical genes is
significantly below this mean length (one-way ANOVA, p<0.01).
The endosymbiont species all showed significantly
lower percentages of uncharacterised genes in their genomes as compared with
the free-living species E. coli and Y. pestis (t-test, p<0.001). This is to be
expected in species that have undergone genome reduction as it is highly likely
such organisms will have minimised both genes that are non-essential and
superfluous layers of redundancy in their genomes.
As mentioned above, a number of predictive methods
were applied to search for characteristic structural features to gain some
tentative insight into the possible functions of the core set of hypothetical
gene products. While caution must be exercised when interpreting the results o
such methods, some interesting possible characteristics of the proteins encoded
by these ORFs were suggested. The results of SignalP signal peptide
prediction, HMMTop topology prediction, PROSITE structural motif
search, and InterPro to scan for known protein family signatures,
domains and functional sites are summarised in Table 3 and Appendix A.
As an example, SignalP predicted the ynfM gene product would have a signal peptide cleaved off around residue 30. PSORT suggested the molecule would be localised to the internal membrane of the bacteria and HMMTop indidcated 12 transmembrane helices with an N-terminal in topology. Both InterPro and SignalP suggested a transport function for this molecule, which would be consistent with its predicted subcellular localisation.
Discussion and Conclusions
With the wealth of molecular and genetic tools
available, the common enterobacterium Escherichia coli is probably the
best understood cell model system. However, elucidation of its genome sequence
yielded the discovery of around 2000 previously uncharacterised open reading
frames (orfs), potentially
encoding products of unknown function (Blattner et al., 1997).
These as yet uncharacterised genes, provisionally given ‘y’-names based on
their map position (Rudd, 1998), account for more than 50% of the E. coli
genome, suggesting that many may be functionally important. However, as these
putative genes have eluded detection by traditional phenotype-based methods for
over a century, it will be difficult to characterise their functions using
standard forward genetics approaches.
One potentially useful approach to investigate the
importance of these ORFs is to search for homologues in closely related species
with reduced genomes.
To establish the relative importance of the
hypothetical E. coli genes, this study was performed to identify which
of these genes have been conserved in related species – Buchnera aphidicola, Candidatus
Blochmannia floridanus, and Wigglesworthia glossinidia – that have
undergone endosymbiotic genome reduction. Such organisms show accelerated loss
of non-essential and redundant genes, implying that any genes retained through
endosymbiotic evolutionary genome degradation and reduction are likely to have
functions essential for survival.
The simple method used here, scanning for matching gene
names and re-checking with blast,
allowed rapid paring of the original ~2000 hypothetical E. coli genes to
a much more focused dataset of genes pre-selected for their central importance
to the cell.
Although rapid, the approach used here does have the
limitation that it relies heavily on accurate and consistent annotation, which
is not always available. Therefore, genes for which the annotations are
inconsistent across strains or species may be missed in the analysis. Indeed, a
number of uncharacterised genes were annotated in such a way as to appear
characterised or were missed in the original screen due to the arbitrary
designations used (‘hypothetical’, ‘putative’, ‘possible’, ‘probable’ etc.),
and annotation-based screening of Y. pestis initially gave the erroneous
impression that none of these genes were conserved in this organism. To control for this, BLAST was used to
identify synonyms with inconsistent annotations as well as for direct
comparisons with the subset of uncharacterised E. coli genes. The ORFs thus identified were then included in the
analyses.
References
- Blattner FR,
Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J,
Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden
MA, Rose DJ, Mau B, Shao Y. 1977.
The complete genome sequence of Escherichia coli K-12. Science Sep
5;277(5331):1453-74. #Ref for E. coli genome sequencing project
- Rudd KE.
1998. Linkage map of Escherichia coli K-12, edition 10: the
physical map. Microbiol Mol Biol Rev. 1998 Sep;62(3):985-1019.
- Andrade et
al. (1999). Bioinformatics 15:391-412
- Iliopoulos
I, Tsoka S, Andrade MA, Janssen P, Audit B, Tramontano A, Valencia A,
Leroy C, Sander C, Ouzounis CA. 2000. Genome sequences and great
expectations. Genome Biology 2000, 2:interactions 0001.1-0001.3 (published
29 December 2000)
- Douglas AE
& Raven JA. 2003. Genomes at the interface between bacteria and
organelles. Philosophical Transactions of the Royal Society of London B
358, 5-17.
- Nielsen H,
Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and
eukaryotic signal peptides and prediction of their cleavage sites. Protein
Engineering 10, 1-6 (1997).
- Nielsen H,
Krogh A. Prediction of signal peptides and signal anchors by a hidden
Markov model. In Proceedings of the Sixth International Conference on
Intelligent Systems for Molecular Biology (ISMB 6), AAAI
Press, Menlo Park, California, pp. 122--130 (1998).
- Tusnády GE,
Simon I. 1998. Principles Governing Amino Acid Composition of Integral
Membrane Proteins: Applications to Topology Prediction. J. Mol. Biol. 283,
489-506.
- Nakai, K.
and Kanehisa, M., Expert system for predicting protein localization sites
in Gram-negative bacteria, PROTEINS: Structure, Function, and Genetics
11, 95-110 (1991).
- Mulder NJ,
Apweiler , Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M,
Bradley P, Bork P, Bucher P, Copley RR, Courcelle E, Das U, Durbin R,
Falquet L, Fleischmann W, Griffiths-Jones S, Haft D, Harte N, Hulo N, Kahn
D, Kanapin A, Krestyaninova M, Lopez R, Letunic I, Lonsdale D,
Silventoinen V, Orchard SE, Pagni M, Peyruc D, Ponting CP, Selengut JD,
Servant F, Sigrist CJA, Vaughan R, Zdobnov EM. 2003. The InterPro Database, 2003 brings
increased coverage and new features.
Nucl. Acids. Res. 31: 315-318. - Tamas I, Klasson L, Canback B, Naslund
AK, Eriksson AS, Wernegreen JJ, Sandstrom JP, Moran NA, Andersson SG. 50 million
years of genomic stasis in endosymbiotic bacteria. Science. 2002 Jun 28;296(5577):2376-9.
- Silva FJ, Latorre A, Moya A. Why are the
genomes of endosymbiotic bacteria so stable? Trends Genet. 2003 Apr;19(4):176-80.
Table 4.
Statistical Analyses
|
Spp |
All
Genes |
Unchar |
Char |
%Unchar |
%Char |
%Unchar/Char |
p
value |
Significance |
|
B.Ap |
619 |
97 |
522 |
15.67% |
84.33% |
18.58% |
|
|
|
B.Bp |
504 |
92 |
412 |
18.25% |
81.75% |
22.33% |
0.754117760734640 |
N/S |
|
B.Sg |
545 |
95 |
450 |
17.43% |
82.57% |
21.11% |
0.850706757429128 |
N/S |
|
|
|
|
|
|
|
|
|
|
|
Wg |
611 |
122 |
489 |
19.97% |
80.03% |
24.95% |
0.340759361807448 |
N/S |
|
C.bloch |
583 |
78 |
505 |
13.38% |
86.62% |
15.45% |
0.763775281188328 |
N/S |
|
|
|
|
|
|
|
|
|
|
|
Y.p |
4090 |
2509 |
1581 |
61.34% |
38.66% |
158.70% |
0.000000000013081 |
SIGNIFICANT |
|
E.coli |
4289 |
2403 |
1886 |
56.03% |
43.97% |
127.41% |
0.000000001161136 |
SIGNIFICANT |
Fig. 6.
ACT Results.
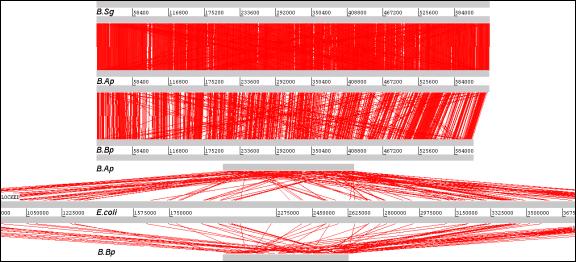
Sequence comparisons were produced by local blastn
searches for analysis using act
(Artemis Comparison Tool). The results indicated extensive synteny between b.ap and b.sg, but a greater degree of chromosomal rearrangement
between b.ap and b.bp. The lower figure shows the
results of comparisons between B.Ap, E. coli K12, and B.Bp.
(figures/Fig6ACT.tif)
Protein: gloB;BU246;probable
hydroxyacylglutathione hydrolase|275795..276550(comp)
Length: 251
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0205
Entropy of the best
path: 17.0206
The best path:
seq MILKKISILS
DNYVWVLLNT SGSCIIIDPG LSEPIIQEIE RKKWRLRAIL
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LTHNHIDHTG
GTRKIIEYFP KISVFGPKET RQHGVNKIVS HGDRIILLDK
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IFYVFFTPGH
TSGHVSYYSQ PYIFCGDTLF SAGCGRVFKN KHLEMYRSIK
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IISSLPDSTL
LCCSHEYTLS NLQFSMFILP NDNFIKLYLK KIEIKLKLGQ
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SSLPSYIFFE
KKINLFLRTN DNYVKKSIGL KSTCTDFEVF KRLRLKKDFW
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq S 251
pred O
Protein:
rnfA;BU113;hypothetical protein|117867..118448
Length: 193
N-terminus: OUT
Number of transmembrane
helices: 6
Transmembrane helices: 4-28
41-65 74-92 103-125 134-154 169-191
Total entropy of the
model: 17.0007
Entropy of the best path: 17.0032
The best path:
seq MKHYILFFIS
NILIENFILV KFLGLCPFLG ASSNIETAFG MSCATTFVIL
50
pred OooHHHHHHH HHHHHHHHHH HHHHHHHHii iiiiiiiiii HHHHHHHHHH
seq TSSVLLWCVN
FFILLPLDLI YLRIIAYMLI VSVSVQFLEI VLRKTSPILY
100
pred HHHHHHHHHH HHHHHooooo oooHHHHHHH HHHHHHHHHH HHiiiiiiii
seq RLLGIFLPLI
TTNCTVLAIP LFSLYEHHTF LESIFYGLSA SLGFALVMII
150
pred iiHHHHHHHH HHHHHHHHHH HHHHHooooo oooHHHHHHH HHHHHHHHHH
seq FSCIRERIVL
SDIPLPFQGA PIILITVSLI SITFMGFKGL IKI
193
pred HHHHiiiiii iiiiiiiiHH HHHHHHHHHH HHHHHHHHHH Hoo
Protein:
ydgO;BU116;hypothetical protein|120631..121629
Length: 332
N-terminus: OUT
Number of transmembrane
helices: 9
Transmembrane helices: 17-41
72-89 100-119 150-169 200-217 224-241 254-271 278-294 303-322
Total entropy of the
model: 17.0096
Entropy of the best
path: 17.0120
The best path:
seq MFLVIVACLP
GIFAKYYFFG IGTLIQIFFS IFISLVLEII ILKIRSKNIK
50
pred Oooooooooo ooooooHHHH HHHHHHHHHH HHHHHHHHHH Hiiiiiiiii
seq NYLQDTSLVL TSVLFGVSIP
PLLPWWMTSI GLFFAIVVAK HLYGGIGQNI 100
pred iiiiiiiiii iiiiiiiiii iHHHHHHHHH HHHHHHHHHo oooooooooH
seq FNPAMVGYAV
LLISFPVYMN NWNERDFSLS FFNDFKKSAY IIFFKNDITT
150
pred HHHHHHHHHH HHHHHHHHHi iiiiiiiiii iiiiiiiiii iiiiiiiiiH
seq VSSSYLNIIP DAFTTATPLN NFKIKSHLKD DFFLKENIIK
NKEVSIQTSW 200
pred HHHHHHHHHH HHHHHHHHHo oooooooooo oooooooooo oooooooooH
seq KCINISFFLG
GIFLLFTKII CWRIPISFLS SLGMLSIITY FYSKELFMSP
250
pred HHHHHHHHHH HHHHHHHiii iiiHHHHHHH HHHHHHHHHH Hooooooooo
seq QVHFFSGGTM
ICAFFIATDP VTAACNNVGK IVFGIIIGFL VWIIRNYSDY
300
pred oooHHHHHHH HHHHHHHHHH HiiiiiiHHH HHHHHHHHHH HHHHoooooo
seq PDAIAFSVLF
ANMTVPLVDY YTKSSGYGRN NI 332
pred ooHHHHHHHH HHHHHHHHHH HHiiiiiiii ii
Protein:
ydgQ;BU118;hypotheical protein|122247..122930
Length: 227
N-terminus: OUT
Number of transmembrane
helices: 6
Transmembrane helices: 34-55
68-86 93-112 127-147 156-175 182-201
Total entropy of the
model: 17.0070
Entropy of the best
path: 17.0098
The best path:
seq MNIKSFLNNR
LWKNNSSLVQ LLGLCPVLAM TTNAINAIGL GMTTTLVLTI
50
pred OOOOOOOOOO OOOOOOOOoo oooooooooo oooHHHHHHH HHHHHHHHHH
seq TNTIISSFRK
IIPKDLRIPI YMMIISSVVT SIEMLLHAYT FNLYQSLGIF
100
pred HHHHHiiiii iiiiiiiHHH HHHHHHHHHH HHHHHHoooo ooHHHHHHHH
seq IPLIVTNCII
VGRADLIAYK SSIVESFFDG IFIGLGSMFA MFAVGSIREI
150
pred HHHHHHHHHH HHiiiiiiii iiiiiiHHHH HHHHHHHHHH HHHHHHHooo
seq LGNGTLFFGA
NKIISNIHSS VFFTLLDKKF TIILAVFPPG GFLILGFLIA
200
pred oooooHHHHH HHHHHHHHHH HHHHHiiiii iHHHHHHHHH HHHHHHHHHH
seq IKNFIDLYYK
KNTIKNIEQC SCSNKIK 227
pred Hooooooooo ooooooOOOO OOOOOOO
Protein: sohB;BU283;possible
protease sohB|308270..309358
Length: 362
N-terminus: IN
Number of transmembrane
helices: 3
Transmembrane helices: 9-31
36-58 190-213
Total entropy of the
model: 17.0164
Entropy of the best
path: 17.0173
The best path:
seq LKSFYIKNYF
FILKNICSIF FKKVYCVNLL LNYELFLAKI ITFIIISISI
50
pred IIIiiiiiHH HHHHHHHHHH HHHHHHHHHH HooooHHHHH HHHHHHHHHH
seq LILFYTIIKR
KKNIQSKIKI TLLQDNYKNV KNKILLSTMK NVEKKIWFKK
100
pred HHHHHHHHii iiiiiiiiii iiiIIIIIII IIIIIIIIII IIIIIIIIII
seq QKEKNKKELL
LKNNKKKLFV LDFKGDVYAN EVVGLREEIS AILLVANKHD
150
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq EVLLRLESSG
GVIHGYGLAA SQLNRLRQKG IRLIVSVDKI AASGGYMMAC
200
pred IIIIIIIIII IIIIIIIIII IIIIiiiiii iiiiiiiiiH HHHHHHHHHH
seq VADYIVSAPF
AIIGSIGVVG QIPNFNKLLK KCNIDFELHT AGDYKRTLTM
250
pred HHHHHHHHHH HHHooooooo ooooooooOO OOOOOOOOOO OOOOOOOOOO
seq FGNNTESTRK
KFCDELNTTH KLFKSFIKEM RPSLDIEDVS NGEHWFGTIA
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LEKKLVDQIG
TSDDILISKM EEYTLLRIQY IYRKKILERF TASVTHNLSE
350
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TLLKIFFYKN YL 362
pred OOOOOOOOOO OO
Protein:
yabC;BU224;hypothetical protein|247278..248216(comp)
Length: 312
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0215
Entropy of the best
path: 17.0215
The best path:
seq MNHIFKHIPV
MKKELIDSLK IKKNGIYIDS TFGTGGHSNE ILKKLGQNGR
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LYSIDRDPIA
FSIGSEIKDS RFHIINENFS KLLDFAKNEK IIGKVNGIIF
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DLGVSSIQID
DYRRGFSFKN DGPLDMRMNP NYGISASEWL FESNVKEISF
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq VLKNFGEERF
SRKIAYAIKR RSQIKKITST LELANIIKKT IPTKNKFKHP
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ARRSFQAIRI
YINQELEEIQ KALESTLKIL KPGGRISIIS FHSLEDRLVK
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KFMIKNSTKA
IIPYGMPITE EQLNRLTTCK LKIINRILPT QNEINNNPRA
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq RSSVLRIAEI QE 312
pred OOOOOOOOOO OO
Protein: yabI;BU139;hypothetical
protein|145105..145875(comp)
Length: 256
N-terminus: IN
Number of transmembrane
helices: 5
Transmembrane helices: 18-42
57-81 140-164 179-196 219-236
Total entropy of the
model: 17.0061
Entropy of the best
path: 17.0082
The best path:
seq MESWLTSFIT
QSLVYSLLVV GIVSFLESLA LVGLLLPGII LMTTLGTFIG
50
pred IIiiiiiiii iiiiiiiHHH HHHHHHHHHH HHHHHHHHHH HHoooooooo
seq DGKLSFYPAW
ISGTTGCLLG DWISYYIGLY FKNWLYNFNF LKKNQKLLDK
100
pred ooooooHHHH HHHHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiiiiIIII
seq TKSFLDKHSM
LTIILGRFIG PTRPLIPMVS GMLKLPLKKF VFPSIIGCIL
150
pred IIIIIIIIII IIIIIIIIII IIIIiiiiii iiiiiiiiiH HHHHHHHHHH
seq WPPVYFFPGI
VTGIAIKIPE SSQSYYFKWL LLLISILIWL GIWLISKWWK
200
pred HHHHHHHHHH HHHHoooooo ooooooooHH HHHHHHHHHH HHHHHHiiii
seq MRKNHIDNRT
SFFTKKKIGW LAILTMSSGI LSLIAIQFHP TMLIFREIFS
250
pred iiiiiiiiii iiiiiiiiHH HHHHHHHHHH HHHHHHoooo oooooooooo
seq TILLGT 256
pred oOOOOO
Protein:
yajC;BU134;hypothetical protein|139801..140136
Length: 111
N-terminus: OUT
Number of transmembrane
helices: 1
Transmembrane helices: 23-40
Total entropy of the
model: 17.0073
Entropy of the best
path: 17.0078
The best path:
seq MSFFIQNANA
VVNGTSESSN SYSLIFMAVI FLLIFYFMLF RPQQKKDKEH
50
pred OOOOOOOooo oooooooooo ooHHHHHHHH HHHHHHHHHH iiiiiiiiii
seq KNLINSLVQG
DEVITTSGLL GRIKKITKNG YILLELNETT EVFIKQDFIV
100
pred iiiiiIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq SLLPKGTLKS L 111
pred IIIIIIIIII I
Protein:
yajR;BU466;hypothetical protein|511376..512548(comp)
Length: 390
N-terminus: IN
Number of transmembrane
helices: 12
Transmembrane helices: 8-30
43-66 75-93 98-122 131-154 159-182 213-231 248-267 276-294 299-321 334-353 366-384
Total entropy of the
model: 16.9955
Entropy of the best
path: 16.9983
The best path:
seq MNFLELQVTL
SFCVIFLLRM LGMFMILPIL SKYGMLLDGG NKFLIGLSMG
50
pred IiiiiiiHHH HHHHHHHHHH HHHHHHHHHH oooooooooo ooHHHHHHHH
seq IYGISQVIFQ
IPFGILSDKF NRKKIILLGL FMFFIGNIIS ASIHSIWGLI
100
pred HHHHHHHHHH HHHHHHiiii iiiiHHHHHH HHHHHHHHHH HHHooooHHH
seq IGRFFQGSGA
ISGVCMAFLS DLIREENRVK SIAAIGVSFA ISFLIAVVSG
150
pred HHHHHHHHHH HHHHHHHHHH HHiiiiiiii HHHHHHHHHH HHHHHHHHHH
seq PIIVHYFGFF
SIFWISAFLS IVCMIIVCFF VPFSKKNILK QNKTLHSYKK
200
pred HHHHooooHH HHHHHHHHHH HHHHHHHHHH HHiiiiiiii iiiiiiiiii
seq VLNFVLNKVF
FRFYLGVFFL HFLLMIKFTM IPNQFEISGF SLDNHWKVYL
250
pred iiiiiiiiii iiHHHHHHHH HHHHHHHHHH Hooooooooo oooooooHHH
seq GTILISFFVL
FLFIFYCKYK YILENIIEIC ILFILFSEII FLSAQKNLLF
300
pred HHHHHHHHHH HHHHHHHiii iiiiiHHHHH HHHHHHHHHH HHHHooooHH
seq LIISLQIFFI
SFNFLEVFLS SHLSRQLSNN YRGSIMSIYS TSQFLGIFFG
350
pred HHHHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiHHHHHHH HHHHHHHHHH
seq GVFSGWLYSF
LNFSQIFYFE LFIILLWLIF SFFLRNRFYL 390
pred HHHooooooo oooooHHHHH HHHHHHHHHH HHHHiiiiii
Protein:
yba2;BU181;hypothetical protein|198321..199037
Length: 238
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0136
Entropy of the best
path: 17.0136
The best path:
seq MTDEEKNLIE
NLFHRLKKTE LNSPERDDAA DELIQRLAKK QPTSSYYMAQ
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TILIQETAIK
KMSIELEELR KKIKILNREE TNKKPSFLSN FFKKKSPSEI
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ISNDNNILKK
KENILPSNYS SSPISPTTQT SPVINNTRSS SFLGNALQTA
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TGVAGGMILG
NMLMNVFSHT KPEEDIFDTV KQSSSDEYTE NNFLNNNTDN
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DLINYSYNES
DINFRESSEE SINNNVDDTD DIDNDNFI 238
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOO
Protein:
ybaB;BU482;hypothetical protein|532636..532965
Length: 109
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0113
Entropy of the best
path: 17.0113
The best path:
seq MFTKGGLGNL
MKQAQQMQEK MAKIQEEIAQ MEVTGEAGAG LVKVTINGAH
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NCRRVEVDPS
LLQDDKDMLE DLAAAAFNDA TRRISEVQKK KMSAISTGMQ
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LPNGFNMPV 109
pred OOOOOOOOO
Protein:
ybeD;BU488;hypothetical protein|538445..538708
Length: 87
N-terminus: IN
Number of transmembrane
helices: 1
Transmembrane helices: 11-28
Total entropy of the
model: 17.0130
Entropy of the best
path: 17.0123
The best path:
seq MKTKLREMLR
FPCFFTYKII GLAQPELIDQ IIKVIQIQIP GDYTPQVKSS
50
pred Iiiiiiiiii HHHHHHHHHH HHHHHHHHoo oooooooooo oooOOOOOOO
seq NRGNYLSVSI
TICAKNFEQI ECLYHEISKI NIVRMVL 87
pred OOOOOOOOOO
OOOOOOOOOO OOOOOOOOOO OOOOOOO
Protein:
ybeY;BU442;hypothetical protein|482486..482821
Length: 111
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0142
Entropy of the best path: 17.0144
The best path:
seq IVDEWEIKKL
NFNYRKKNKP TNILSFPFNK FIKINYKLLG DLVLCKNIIE
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KESLKYNKSL
ESHWAHITIH GTLHLLGYDH QNNKEADIME RLENKIMLSL
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NYKKPHILKS F 111
pred OOOOOOOOOO O
Protein:
ybgI;BU301;hypothetical protein|332275..333018
Length: 247
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0202
Entropy of the best
path: 17.0202
The best path:
seq MNNFLLEDII
NKKLLSNQYQ DTVPNGLQIE GTEIVKKIIT GVTACQALLD
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KALFYNADTL
IVHHGYFWKN ESKYIHNMQR QRLKTILSHN INLYSWHLPL
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DVHPKLGNNA
QIAKKLNIDI QGSILPYVLW GTTKNKMTGF EFANKIERKF
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KKYPIHLYEN
APLYISRVAW CSGRGQGFIK KACAFGIDAF LTGEISEETT
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq HIAKELGIHF
FSLGHHATEK DGVKSLGEWL QRKYDLCVDF IDIYNPA
247
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOO
Protein:
ybhE;BU293;hypothetical protein|321520..322524(comp)
Length: 334
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0293
Entropy of the best
path: 17.0293
The best path:
seq MKQVVYIANS ESKNIEVWNL
CKSGKMNLIQ KIETDGKIQP INIIQKRNLL 50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq YAGIFPDNKI
ITYSINHNGF LEKKNESNIP GKANYISFDK KKEFLFCSSY
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq HSNFISVSPL NKFGIPQNPI QIIYNIEGCH AAKMNYKYNI
LFVISLKEDC 150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IYLYYLTDFG
ILKSTEQNIL HTQKKSGPRH IIFHPNQDFI YTINELNGTI
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DVWKIYKKNN
VIKVKNIQNI HVLKNRFLKD YWCSDIHITS CGRFLYACDR
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq FFNIISLFHI
NQNDNKLVFF KSYDTEEQPR SFNINSHNTH LIVAGEKSNT
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq FIIYSISNST
GELKKINVYS TGQRPVWILI HALC 334
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOO
Protein:
yccK;BU467;hypothetical protein|512622..512966
Length: 114
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0141
Entropy of the best
path: 17.0141
The best path:
seq MNTDNKILYT
YENLEKDSEG YLKKTKDWNI KLAEEIAKRE NITLSSDHWK
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq VIIFVRKFYF
KFNITPSMRM LIKSIQKEIG KSKMNSIYLF KLFPKGPAKQ
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ASKIAGIPKP
VKCL 114
pred OOOOOOOOOO OOOO
Protein:
yceA;BU365;hypothetical protein|400570..401544(comp)
Length: 324
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0301
Entropy of the best
path: 17.0301
The best path:
seq MSILHNIVSK
KELKRRMFFE TEPRLTLSFY KYFFIKNTQE YRDRLYKTFY
50
pred OOOOOOOOOO
OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KYNVLGRIYV
ASEGINAQIS VPKKYYSILK KFLYNFDIEL NNLRINKSLD
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NEKSFWVLCV
KIKKKIVQDG IKEHFFNPNN VGIYIQSEQV NSMLNDKKTI
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq FIDMRNSYEY
AIGHFENAIE IKSITFREQL KKVIQLMAYA KNKKIVMYCT
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GGIRCEKATS
WMLFNGFKHV YHLEGGIIGY VHDARKNGLP VLFKGKSFVF
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DNRMSEKISD
EVISYCKQCG KSSDVYINCK YSSCHLLFIQ CENCSVKFHS
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq CCSLECMKKY
KFYMLNNDLK KISY 324
pred OOOOOOOOOO OOOOOOOOOO OOOO
Protein:
ycfF;BU357;hypothetical protein|389976..390320
Length: 114
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0166
Entropy of the best
path: 17.0166
The best path:
seq MQDNSIFKNI
IQRKIPANIV YQDKKITAFE DIKPKAPVHI LIIPNFFISS
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SNDINKKNKW
IMSHMFYIAV KIAKQKKINQ EGYRIIINCN EYGGQEINYL
100
pred OOOOOOOOOO
OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq HMHLLGGKKL
KSFS 114
pred OOOOOOOOOO OOOO
Protein:
ycfH;BU355;hypothetical protein|387632..388426
Length: 264
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0201
Entropy of the best
path: 17.0201
The best path:
seq MFLIDSHCHL
DRLNYNLPLE NIEDVLKKSY QNHVKNFLTV STCISNFYNI
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KKLFKKYNTI
FYSCGVHPLN CKKELNLFHT IENLSNEIKK LSCIKDVIAL
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GETGLDYYYS
SDTKKIQQDF FREHIRVAIK LKKPIIVHSR NASEDTIKIL
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QEENAEKCKG
VLHSFTGDYN TACKLLDLGF YISCSGIITF KNSLELCKTI
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq RKIPLNRLLI
ETDSPYLSPA PYRGKGNQPA YLFYIAEYLS ILKEIDIHAL
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GHITTSNFRT
LFNI 264
pred OOOOOOOOOO OOOO
Protein:
ychE;BU267;hypothetical protein|293524..294171
Length: 215
N-terminus: OUT
Number of transmembrane
helices: 6
Transmembrane helices: 8-31
52-76 81-100 121-140 149-168 189-213
Total entropy of the
model: 17.0001
Entropy of the best
path: 17.0020
The best path:
seq MNISIFDLSI
YIKFFIGLCA LVNPIGMIPI FTTMTNNQSF LERKKTNIVA
50
pred OooooooHHH HHHHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiiiiiiii
seq NFSVSLILLI
SLFFGSNILN IFGISINSFR IAGGILIISI AFSMISGQFI
100
pred iHHHHHHHHH HHHHHHHHHH HHHHHHoooo HHHHHHHHHH HHHHHHHHHH
seq KTIKTKKETK
EENKIDNISV VPLAMPLIAG PGAISSTIVW STYYSSWANL
150
pred iiiiiiiiii iiiiiiiiii HHHHHHHHHH HHHHHHHHHH ooooooooHH
seq FLCSLVIFLF
SFVCWLCFEA APYVVQILGN TGINIITRIM GLLLMSLGIE
200
pred HHHHHHHHHH HHHHHHHHii iiiiiiiiii iiiiiiiiHH HHHHHHHHHH
seq FISTGIGAIF
PGLLH 215
pred HHHHHHHHHH HHHoo
Protein: ychF;BU191;probable
GTP-binding protein|205836..206924
Length: 362
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0241
Entropy of the best
path: 17.0240
The best path:
seq MGFKCGIIGL
PNVGKSTLFN LLTKGNSAVA NFPFCTIKPN IGIVPVIDER
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq INNLNQIVSP
QKTVNAFIEF IDIAGLVKGA SQGEGLGNQF LGNIRDVHAI
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq AHVVRCFKDD
NITHIYNQVQ PIKDIDIINS ELILSDFDLC EKTILKLQKK
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TLLKNKETQE
KINTLKKCLN HLKQFFMLKT LNLNKTEKQL ISYLRFLTLK
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq PTMYIANINE EKESYYFLDK
LNEIAKKEGS IVIPIHANLE LDLVKMSDEE 250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KKSFMKLFNI
KTLGLNSIIS SGYHLLNLIT FFTVGDKEIR AWAIPNGSTS
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IEAAHKIHSD
FSKGFIRAQI IKYVDFITYK SEAKIKEMGK FRTEGKQYYI
350
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QDGDIIHFLF NV 362
pred OOOOOOOOOO OO
Protein:
yciA;BU274;hypothetical protein|300079..300486(comp)
Length: 135
N-terminus: IN
Number of transmembrane
helices: 3
Transmembrane helices: 29-46
61-78 103-120
Total entropy of the
model: 17.0129
Entropy of the best
path: 17.0128
The best path:
seq MSEKNKLPKG
IIVLKTLSMP ENINANGDIF GGWIMSQMDL GGAILAKEIS 50
pred IIIIIIIIII IIIiiiiiii iiiiiiiiHH HHHHHHHHHH HHHHHHoooo
seq GGKVATVRVD
SINFLKSVSV GDIVNCYANC IKIGKSSIKI NVEIWIKKIY
100
pred oooooooooo HHHHHHHHHH HHHHHHHHii iiiiiiiiii iiiiiiiiii
seq SKPLGQYYCA
AEAIFIYVAI NKTGQPRELL PMSII 135
pred iiHHHHHHHH HHHHHHHHHH oooooooooo ooooo
Protein:
yciC;BU276;hypothetical protein|301084..301827(comp)
Length: 247
N-terminus: IN
Number of transmembrane
helices: 6
Transmembrane helices: 20-41
88-107 120-139 144-166 189-208 217-238
Total entropy of the
model: 17.0045
Entropy of the best
path: 17.0059
The best path:
seq MPITVNKLRH
DTHHFFYKKI GAIFFISIFA TFMNILIDMF IKPDMHIVSI
50
pred IIIIiiiiii iiiiiiiiiH HHHHHHHHHH HHHHHHHHHH Hooooooooo
seq MENNKFINTS
SLLEFIQNMN LNEKHELLKY SILKIMESLI SKTTLLGSII
100
pred ooooooOOOO OOOOOOOOOO OOoooooooo oooooooHHH HHHHHHHHHH
seq ILISVVSEPK
KKSIVSSIRT FFLFFPSLFI LNFLTTFIIQ IGFMLLIIPG
150
pred HHHHHHHiii iiiiiiiiiH HHHHHHHHHH HHHHHHHHHo oooHHHHHHH
seq ILLSIILSLS
PIILFFKKNR LLDSIRLSMY ISWKYIKIIG PGVLFWMCGK
200
pred HHHHHHHHHH HHHHHHiiii iiiiiiiiii iiiiiiiiHH HHHHHHHHHH
seq FILTMLLAHF
SLINKNVLFL ISNISMNILF SILIIYLFRF YMIFLRS
247
pred HHHHHHHHoo ooooooHHHH HHHHHHHHHH HHHHHHHHii iiiiiii
Protein:
ydiC;BU122;hypothetical protein|127212..127604(comp)
Length: 130
N-terminus: OUT
Number of transmembrane
helices: 2
Transmembrane helices: 29-46
77-94
Total entropy of the
model: 17.0144
Entropy of the best
path: 17.0144
The best path:
seq MNKNEVKTYL
LKKNTFQNIS ITKDAIEQIL FLINLNSDNI GIRLSIKKSG
50
pred OOOOOOOOOO OOOooooooo ooooooooHH HHHHHHHHHH HHHHHHiiii
seq CAGFRYSMKL
LKASELKKEK DEKEVSFFYQ NILIYIYSKD IPFLEGIRID
100
pred iiiiiiiiii iiiiiiiiii iiiiiiHHHH HHHHHHHHHH HHHHoooooo
seq FVKNNINKIF
KFYNTKLEKF CGCGESFSIN 130
pred oooooooooO OOOOOOOOOO OOOOOOOOOO
Protein:
yeaZ;BU324;hypothetical protein|360644..361309
Length: 221
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0187
Entropy of the best
path: 17.0188
The best path:
seq MSNIILSIES
SLDCCSVAIY KNEYIHSLSE KCKKKHTTHI LPMIKEILSQ
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TKTEFKELNY VSFSKGPGNF
TSIRIAASIA QSLSISLKIP IISVSTLAIM 100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq AEKTFRKYKQ
KDVIVAIHAK KKQVYWAKYT RNKNSIWIGE YTESLLEKKI
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IQEKIENLKK
RWTLVSDQSE LIEFQNILNV KKNYVFLPNA KDIIPFVLLK
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IKNKNKSFSI
ENNINYLYNQ F 221
pred OOOOOOOOOO OOOOOOOOOO O
Protein:
yfgB;BU286;hypothetical protein|313130..314221
Length: 363
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0254
Entropy of the best
path: 17.0254
The best path:
seq MNNHIDIFNI
PISKINLLDL NRQNLKYFLI SLGAKNFCTE QVMSWIYNYY
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq CDDFNKMLNI
SIKTRKKLYE KSYIFASEFI EEKISYDGTI KWITDINNQK
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IETVYMPEKK
RSTLCVSSQI GCSLKCHFCA TGQEGFQRNL KVSEIIAQIW
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QANKRLKEKN
IKKNITNIVF MGMGEPLLNL KNVVSALTII LDEYGFGLSK
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq RRVTLSTSGI
VPALDKLRNM IDVSLAISLH APNDFIRNII MPINRKYNIS
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SVLSSALKYF
KYSNANRGGI TIEYVMLDRI NDSNENARQL SVLLSKIPSK
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq INLIPWNSFS
GPSFLCSNTD RINMFANILR KKGFTTTIRK NRGEDINAAC
350
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GQLTGIITNY
FKK 363
pred OOOOOOOOOO OOO
Protein:
yfgM;BU608;hypothetical protein|639860..640441(comp)
Length: 193
N-terminus: OUT
Number of transmembrane
helices: 2
Transmembrane helices: 9-32
63-80
Total entropy of the
model: 17.0139
Entropy of the best
path: 17.0143
The best path:
seq MLNISKKNII
FFILFFLIIS LILFNWKYFS LVNKENLESL KYEKIIKKIN
50
pred OOOOOOooHH HHHHHHHHHH HHHHHHHHHH HHiiiiiiii iiiiiiiiii
seq KKKSKNLYEV
ENFIVQNTSI YGTLTALSLA KKYVECNNLD KALLQLNNSL
100
pred iiiiiiiiii iiHHHHHHHH HHHHHHHHHH oooooooooo oooooOOOOO
seq KYTKEENLKN
LLKINIAKIQ IQKNENNKAM NILETIQNHN WKNIIEHMKG
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DIFININNKK
EAIKSWKKSL FIEDSNASKE IINMKLNELK EQN
193
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOO
Protein:
yfhC;BU255;hypothetical protein yfhC|280870..281355
Length: 161
N-terminus: OUT
Number of transmembrane
helices: 2
Transmembrane helices: 26-43
74-93
Total entropy of the
model: 17.0177
Entropy of the best
path: 17.0185
The best path:
seq MKYEKDKNWM
KIALKYAYYA KEKGEIPIGA ILVFKERIIG IGWNSSISKN
50
pred OOOOOOOOOO oooooooooo oooooHHHHH HHHHHHHHHH HHHiiiiiii
seq DPTAHAEIIA
LRGAGKKIKN YRLLNTTLYV TLQPCIMCCG AIIQSRIKRL
100
pred iiiiiiiiii iiiiiiiiii iiiHHHHHHH HHHHHHHHHH HHHooooooo
seq VFGANCNSSD
HRFSLKNLFC DPQKDYKLDI KKNVMQRECS DILINFFQKK
150
pred ooooooooOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq RKNKIHICKK I 161
pred OOOOOOOOOO O
Protein:
yfjF;BU253;hypothetical protein|279980..280279(comp)
Length: 99
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0131
Entropy of the best
path: 17.0131
The best path:
seq MKIIKVTVVY
ALPKIQYICQ VDIALGSTVK DAILKSNLLN LTNDVSFHHN
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq RIGIYNKTVH
LKFKIKDGDR IEIYRNLTID PKEWRRNNVF LSKKLKKIY
99
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOO
Protein:
ygfZ;BU435;hypothetical protein|474156..475115(comp)
Length: 319
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0252
Entropy of the best
path: 17.0252
The best path:
seq MPSFISIQNI
IYPSNELSLT MILLEEWSLT YVEGIDSKKY LQGQLTIDIN
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LLLKTHHTLC
AHCNFNGRVW STMHLFHYEK GYAYIQRKSV SQIQIKEICK
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq YSIFSKIKIR
ELNSICLIGF AGCNVRSFLS SLFVKIPNQS CPVIHEDNKT
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ILWYEKPSER
FLLVLPFLDF LTLKRKINQN IFLNNSKQWL LLDIEAGLPV
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IDKICSNKFT
PQAINLHNLK AISFKKGCYY GQETIARIFF KKTNKYFLCF
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LVSTGSIFPK
IGSFIETKVD SEWFKVGVLL SIVHVKCEEI YIQVVLRKSV
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NINNLFRIHG
FENIFLIKN 319
pred OOOOOOOOOO OOOOOOOOO
Protein:
yggB;BU452;hypothetical protein|493043..493960
Length: 305
N-terminus: OUT
Number of transmembrane
helices: 4
Transmembrane helices: 51-68
89-111 120-143 162-179
Total entropy of the
model: 17.0127
Entropy of the best
path: 17.0147
The best path:
seq ILFIKIEIIA
KTLLIINKKY TEIKMDELNV VNNINHAGTW LIRNQELLLR
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOooooo oooooooooo
seq YTINLTSAII
ILVVGMFISK IISNGANQVL ITRNIDATIA GFLSALMRYI
100
pred HHHHHHHHHH HHHHHHHHii iiiiiiiiii iiiiiiiiHH HHHHHHHHHH
seq IITFTFIAAL
GRIGVQTTSV IAILGAAGMA IGLALQGSLS NFAAGVLLVT
150
pred HHHHHHHHHH HooooooooH HHHHHHHHHH HHHHHHHHHH HHHiiiiiii
seq LRPLKTEEYV
DLGSVSGTVL NIHIFYTTLR TLDGKIVVVP NNKIISGNII
200
pred iiiiiiiiii iHHHHHHHHH HHHHHHHHHo oooooooooo ooooOOOOOO
seq NYSREPARRN
EFIISVSYNS DIDLVIKILR SVIEKEERVI KDKDIIVGLS
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ELAPSSLNFI
VRCWSKNHDL NTVYWDLMAK FKKELDKNNI NIPFPQLDVH
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq VYKKK 305
pred OOOOO
Protein:
yggH;BU551;hypothetical protein|586987..587706(comp)
Length: 239
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0208
Entropy of the best
path: 17.0208
The best path:
seq MKNNIITPRY
NLEGVFLRQI HSFVCRKGRT TTSQLSAIKK YWSLIGVDFQ
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LNALNFSSIF
KHRAPIILEI GFGSGESLVK TAMNFPEKNF LGIEVYKSGI
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GSCLHYASSY
QIQNLRIIYY DATEVMYNMI PDDTLSKVQI FFPDPWHKKR
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq HHKRRLLKNI
FLKIITKKLI IDGILHIATD SESYAFYILD EIKDIKNYKN
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LSEKNNFVKR
PVSRIITKFE KKGLLQGKKI FDLMFQLKK 239
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOO
Protein:
yggS;BU549;hypothetical protein|585205..585807
Length: 200
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0167
Entropy of the best
path: 17.0167
The best path:
seq LKKITIIAVS KNRNINNIEE
AIRSGINNFG ENYLQESLIK IENLKKYKNI 50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TWHFIGKIQS
NKTKKIAQNF SWCQTVDREK IAVLLNKFRP KNLPPINVLI
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QINNLKELQN NRYIDQYQEL AQLILSMPNL NLRGIMAVPS
IKTNVIENNL 150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QYEKIKTIFN
RFKRQYSSVD TLSLGTSVDI KESLLATSNM VRIGRNIFNI
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
Protein:
yggW;BU550;hypothetical protein|585850..586980
Length: 376
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0292
Entropy of the best
path: 17.0292
The best path:
seq MFTLPPISLY IHIPWCLKKC GYCDFYSYVS KEIIPENKYI
EHLLRDFERD 50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LSLINNRNIN
TIFIGGGTPS LLKNTSIKNL LNGIKKRKII SKNIEISIEA
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NPKTLEYQNF
IQYKNSGINR FSLGIQTFNS KMLKKIERTY NSKDAMNAII
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ESKKISDNIN
LDLMYGLPGQ SLEEALSDLQ IAIQCNPSHI SWYQLTIEPN
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TVFYAKKIQT
PHQDVVFNML IEGDKLLKKA GYKKYEISSY SKFNYQCQHN
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LNYWNFGDYI
GIGCGSHGKI TQKNGEIIRT IKNKNINDFL SGKYINSVYQ
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq VSKRDKIFEY
FMNVFRLYKP IFKKHFRENT NIEESFIEKN IQIAIQEGFL
350
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq INQSDCWHTT
KKGKNFLNSL LEIFLK 376
pred OOOOOOOOOO OOOOOOOOOO OOOOOO
Protein:
yggX;BU553;hypothetical protein|588828..589109
Length: 93
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0150
Entropy of the best
path: 17.0150
The best path:
seq LKILKKIFFK
KRSIKIMNRI IFCTFFKKKS EGQDFQSYPG KLGKKIYDQI
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SKKAWEKWIE
KQTILINEEN LNMFNLEHRK KIEKYMKLFL FKK
93
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOO
Protein:
yheL;BU530;hypothetical protein|565030..565302(comp)
Length: 90
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0122
Entropy of the best
path: 17.0123
The best path:
seq MKSPFETNVS
LVISMLKKSD DFLALQDGVL IALKDNIFLK SIIMSPVKLY
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LIKEDVYARG
IRKNISREFI LINYIHFVSL TLKHKKQMTW 90
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
Protein: yheM;BU531;hypothetical
protein|565329..565688(comp)
Length: 119
N-terminus: IN
Number of transmembrane
helices: 1
Transmembrane helices: 21-40
Total entropy of the
model: 17.0126
Entropy of the best
path: 17.0135
The best path:
seq MKMVAFVFSH APHGISLGRE
GLDAIFSISS IFKKISVFFI GDGVLQLIKN 50
pred IIIIIiiiii iiiiiiiiii HHHHHHHHHH HHHHHHHHHH oooooooooo
seq QQPEHILARN
YTSSFSILSL YNIKDLYCCK ASLLERGLNN NNNFILNIDV
100
pred oooooOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LDSYNLRLKL DNYDAIINF 119
pred OOOOOOOOOO OOOOOOOOO
Protein:
yheN;BU532;hypothetical protein|565707..566093(comp)
Length: 128
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0132
Entropy of the best
path: 17.0132
The best path:
seq MNYTILVTGP
PYGTQNSSTA FLFCQSLLKT KHILHSVFFY CDGVLNANNM
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq TTPAIDEFNL
INAWQGLNKK HQVKLYVCNS AALRRGVIED EKLFNMNVKK
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GNLALSFQLS
GLIELAKSIK ICDRIIQF 128
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOO
Protein:
yhfC;BU535;hypothetical protein|568906..570072
Length: 388
N-terminus: IN
Number of transmembrane
helices: 12
Transmembrane helices: 7-31
48-67 76-98 103-122 131-155 160-181 206-227 244-263 272-290 299-320 329-348
357-380
Total entropy of the
model: 16.9934
Entropy of the best
path: 16.9968
The best path:
seq MTNINRIGLT
WISFLSYAFT GALVVVTGMI MGNISNYFHL SISQMSNIFT
50
pred IiiiiiHHHH HHHHHHHHHH HHHHHHHHHH Hooooooooo oooooooHHH
seq FLNAGILVSI
FINSWLIEII SLKKQLIFSF ILTIIAVIGI VLCNSIFLFS
100
pred HHHHHHHHHH HHHHHHHiii iiiiiHHHHH HHHHHHHHHH HHHHHHHHoo
seq INMFILGLVS
GITMSIGTFI ITHLYSGSKR GSLLLLTDSF FSMSGMIFPI
150
pred ooHHHHHHHH HHHHHHHHHH HHiiiiiiii HHHHHHHHHH HHHHHHHHHH
seq VTAYLLEKKI
IWYWSYICIG AIYLLIFLLT INSSFEKFKT NTKNSKETKE
200
pred HHHHHooooH HHHHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiiiiiiii
seq KWNFNVFLLS
ISALLYILGQ LGFISWVPQY ATEIMNIDIK KTGSLVSGFW
250
pred iiiiiHHHHH HHHHHHHHHH HHHHHHHooo oooooooooo oooHHHHHHH
seq MSYMLGMWFF
SFIIKFFNLY RMFIFLTSMS TILMYCFIKS ENFLNQQYII
300
pred HHHHHHHHHH HHHiiiiiii iHHHHHHHHH HHHHHHHHHH ooooooooHH
seq ISLGFFSSAI
YTIIITLASL QTKHPSPKLI NLILLFGTIG TFLTFIITSP
350
pred HHHHHHHHHH HHHHHHHHHH iiiiiiiiHH HHHHHHHHHH HHHHHHHHoo
seq IVEAKGLYVT
LISSNILYGI VFFLSILIYF NKKYERVI 388
pred ooooooHHHH HHHHHHHHHH HHHHHHHHHH iiiiiiii
Protein:
yhgI;BU544;hypothetical protein|579687..580265(comp)
Length: 192
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0147
Entropy of the best
path: 17.0147
The best path:
seq MINISKKAQE
HFTSLLSNEP ENTQIRVFIV NPGTPNAECG VAFCPENEIE
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq LSDIQLKYDG
FFVYVNKDTI SYLKNSVIDL VTDKIGSQLT LKAPYAKNNF
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SKKVSSSLEE
KVKCFLNLEI NPQLSMHGGR VELIKIDKNG IAAIQFSGGC
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NGCSMIGSTL
KETVEKKLLS SFSEIKKVYD ETHHLHGQHS FY 192
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OO
Protein:
yhgN;BU449;hypothetical protein|490053..490601
Length: 182
N-terminus: IN
Number of transmembrane
helices: 5
Transmembrane helices: 24-43
58-77 92-111 126-145 160-179
Total entropy of the
model: 17.0014
Entropy of the best
path: 17.0020
The best path:
seq MDPLGNLPIF
MTILKHLDVK RRRIVVIREM IIALIVMLLF LFVGEKILII
50
pred IIIIIIIIii iiiiiiiiii iiiHHHHHHH HHHHHHHHHH HHHooooooo
seq LNLKTETVSI
SGGVILFLIA IKMIFPSEDN NNEISSSEEP FLVPLAIPLV
100
pred oooooooHHH
HHHHHHHHHH HHHHHHHiii iiiiiiiiii iHHHHHHHHH
seq AGPSLLATLM
LLSHQYLHHM FYLVGSLLIS WFFTVIILLS SSLFLKLFGS
150
pred HHHHHHHHHH Hooooooooo oooooHHHHH HHHHHHHHHH HHHHHiiiii
seq KGVNALERLM
GLVLIMLSTQ MFLDGIRAWF KN 182
pred iiiiiiiiiH
HHHHHHHHHH HHHHHHHHHo oo
Protein:
yibN;BU052;hypothetical protein|56934..57368
Length: 144
N-terminus: OUT
Number of transmembrane
helices: 2
Transmembrane helices: 12-29
94-111
Total entropy of the
model: 17.0118
Entropy of the best
path: 17.0124
The best path:
seq MQDVIFFLSK
HILLISIWIF CFIAAVFFIT RTLLSKSKMI NNFQAIKLIN
50
pred OOoooooooo oHHHHHHHHH HHHHHHHHHi iiiiiiiiii iiiiIIIIII
seq QDKAIVVDTR
SLESFKEGHI LNSINVPLKN IFLGKIKEIE IYKMFPIILV
100
pred IIIIIIIIII IIIIIIIIII IIIIIIIIii iiiiiiiiii iiiHHHHHHH
seq LSDTYKVNAC
IKKFFEYGFN RVYILKNGLY YWKTDNLPLI VNDK
144
pred HHHHHHHHHH Hooooooooo ooooooOOOO OOOOOOOOOO OOOO
Protein:
yigL;BU028;hypothetical protein|29403..29972
Length: 189
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0193
Entropy of the best
path: 17.0193
The best path:
seq MYRIIAVDLD
GTLLTSENKI TKYTKEIIQI LIQKKFYFVF ASGRHYIDIM
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KIKDSLKINI
FIISSNGSKI YNLDNNLIFS DNLDENIASK LCRIKYSDKE
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IITQVYQNDQ
WYINNNKVEN NFCSLLSSLQ YKYFYPDDLN FKNISKIFFT
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SRNFQKLHIL
KRKIINFYGN KVHVNFSIPG CLEIVSGTT 189
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOO
Protein:
yleA;BU441;hypothetical protein|481012..482331
Length: 439
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0280
Entropy of the best
path: 17.0280
The best path:
seq MKYIYIKTWG
CQMNEYDSSM IITLLEKNNQ YSLTKSAENA DILILNTCSI
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq REKAQEKVFH
QLGRWKKIKN NNPKVIIAVG GCVATQEGKE IFKRANYVDI
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq IFGTQTLHRL
PKMIDEVEKK RKLSIDISFP KLEKFKYFLA PKKKGYTADI
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq SIMEGCNKYC
SFCVVPYTRG NEISRPCDDV LFEISLLAKQ GIKEINLLGQ
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NVNAYQGPTF
NGKVCYFSEL IRLVAEIDGI ERIRFTTSNP LEFTDDIIEV
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq YKDTPKLVSF
LHLPVQSGSN KILNLMKRSY TTEDYTSIIK KLTIARPDIQ
300
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq ISSDFIVGFP
GESEIDFEKT IEFIKNINFD MSFSFIYSAR PGTPASNMND
350
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DLDLKEKKRR
LYILQERINI QTMLWSRKMF GSIQSVLVEG VSDKNIMDLY
400
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq GRTENNRVVT FQGSSKMIGQ
FVNVKIKKVH THSLKGELF 439
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOO
Protein:
ynfM;BU588;hypothetical protein|621227..622459(comp)
Length: 410
N-terminus: IN
Number of transmembrane
helices: 12
Transmembrane helices: 27-51
64-83 92-116 121-138 147-171 176-199 228-247 264-281 294-310 315-339 352-375
380-398
Total entropy of the
model: 16.9954
Entropy of the best
path: 17.0016
The best path:
seq LTLLKNKKQL
LEKQYIKKNT KKFNQVILAL FSGGFATFSI LYCVQSILPM
50
pred IIIIIIIIII Iiiiiiiiii iiiiiiHHHH HHHHHHHHHH HHHHHHHHHH
seq FSKQFYLTPA
ESSLALSAAT ITMSLGMLFT GPLSDIIGRK SIMSTSLFIA
100
pred Hooooooooo oooHHHHHHH HHHHHHHHHH HHHiiiiiii iHHHHHHHHH
seq AMLTMICSMM
TSWISIVLLR ALTGLALSGV VAVAMTYISE EIHPNSLSFC
150
pred HHHHHHHHHH
HHHHHHoooo HHHHHHHHHH HHHHHHHHii iiiiiiHHHH
seq MGLYISGNTI
GGFLGRLLSS ILAEKFSWSI SLMVIGLFSF ISSCFFLYFL
200
pred HHHHHHHHHH HHHHHHHHHH HooooHHHHH HHHHHHHHHH HHHHHHHHHi
seq PPSKNFLSVS
INFHKCLHRF YLQLKNRVLF FLFIIGFILM GSFVTIFNYI
250
pred iiiiiiiiii iiiiiiiiii iiiiiiiHHH HHHHHHHHHH HHHHHHHooo
seq GYRLMLEPFF
LCQSSIGLLS TIYLTGVYSS PKAGVLINKY NRNNILIVSL
300
pred oooooooooo oooHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiHHHHHHH
seq MLMIIGLFIT
QYNQLFIIIL GLIIFSGGFF ASHSTASSWV GSYSNIAKIQ
350
pred HHHHHHHHHH ooooHHHHHH HHHHHHHHHH HHHHHHHHHi iiiiiiiiii
seq ATSLYLFFYY
LGSSVFGTFG GFFWFHMQWL GISVFIITML FFGVFLSFKL
400
pred iHHHHHHHHH HHHHHHHHHH HHHHHooooH HHHHHHHHHH HHHHHHHHii
seq KQKNFKNKYF 410
pred iiiiiiiiii
Protein:
yoaE;BU323;hypothetical protein|359049..360614
Length: 521
N-terminus: OUT
Number of transmembrane
helices: 7
Transmembrane helices: 12-35
48-71 80-97 122-145 154-171 184-201 210-227
Total entropy of the model: 17.0087
Entropy of the best
path: 17.0093
The best path:
seq MEFFLDPSTW
AGLLTLVILE VVLGIDNLIF VAILSEKLPP NQRDKARLIG
50
pred OOoooooooo oHHHHHHHHH HHHHHHHHHH HHHHHiiiii iiiiiiiHHH
seq LGLALVMRLA
LLSLISWIVT LNSPIVHNKF FSLSIRDIIL LFGGFFLLFK
100
pred HHHHHHHHHH HHHHHHHHHH HooooooooH HHHHHHHHHH HHHHHHHiii
seq TTMELHERLE
NNHHENSENK NYAGFWAVVI QIVVLDAVFS LDAIITAVGM
150
pred iiiiiiiiii iiiiiiiiii iHHHHHHHHH HHHHHHHHHH HHHHHooooo
seq VNQLLIMMIA
VILATFLMLL ASKALTNFIN LHQTVVVLCL SFLLMIGFSL
200
pred oooHHHHHHH HHHHHHHHHH Hiiiiiiiii iiiHHHHHHH HHHHHHHHHH
seq VTEALRFCIP
KGYLYAAIGF SILIEIFNQI ARHNFMKNQS RRPMRQRAAE
250
pred HooooooooH HHHHHHHHHH HHHHHHHiii iiiiiiiiii iiIIIIIIII
seq AILRLMVGEQ
NKKQQIKKIE INSQKTDSIQ SSKEMETFKD EERYMINGVL
300
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq TLAGRSIRSI
MTPRSNISWV NTEKNTDEIR MQLLDTPHSL FPVCKGELDE
350
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq IIGIVRAKEL
LVAIEKKIDA STFSSKILPI IIPDTLDPIK LLGVLRRAQG
400
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq SFVIVSNEFG
VVQGLITPLD VLEAIAGEFP DADETPDIIQ ENNSWLVKGE
450
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq TDLHSLQQLL
NTEELIKEDN YASLGGLLIA QKGQLPIPGE IIHIHPFYFH
500
pred IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII IIIIIIIIII
seq IVKATEYRID
LVRIIKNQDD N 521
pred IIIIIIIIII IIIIIIIIII I
Protein: yqgF;BU548;hypothetical
protein|584680..585087
Length: 135
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0148
Entropy of the best
path: 17.0148
The best path:
seq MIVIAFDFGI
KKIGVAVGEN ITKKGRPLSV LNAQNGCPNW QLVKNLIQYW
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq QPQFIVVGLP
LNINGTKQKI TNKSEKFANL LKYKFNIVVK MHDERLTTVE
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq AKSIIFKKNG
FKGLKEEKIH SCAAVIILES WFNQY 135
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOO
Protein:
yraL;BU091;hypotheical protein|93393..94241(comp)
Length: 282
N-terminus: OUT
Number of transmembrane
helices: 2
Transmembrane helices:
82-101 112-130
Total entropy of the
model: 17.0218
Entropy of the best
path: 17.0222
The best path:
seq MNEFYIGILY
IVPTPIGNLS DITYRALEVL KDVDIIAAEN IRHTNILLQH
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq FNIKNNLILM
NKDNEKKQSH NLIQELKKGK KIALVSNAGT PIINDPGCIL
100
pred OOOOOOOOOO OOOOOOoooo oooooooooo oHHHHHHHHH HHHHHHHHHH
seq IKQCHIFDIK
VIPLPGACAA ITALSASGII NNRFCYEGFL PSRKKSRCDL
150
pred Hiiiiiiiii iHHHHHHHHH HHHHHHHHHH oooooooooo oooooOOOOO
seq LHSLKEETRT
IIFYESKHRI LESIKDIIEQ IDKNRHIVIA REMTKKWESI
200
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq YGAKASLILE
WLKENKYRYK GEMVIIIDGF KKLKNYTLSK KILDTFSILR
250
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq KFFSLKTSVL
ITAQIHDINK NKLYQYVIKK EE 282
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OO
Protein:
yrbA;BU385;hypothetical protein|425949..426191
Length: 80
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0125
Entropy of the best
path: 17.0125
The best path:
seq MDNQEIKLLL
IKKLNLEQAN ITGDSNHIKI IAIGNIFKNV SQVKRQQIIY
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq APLIDMIKEK
HIHAVSIMSY TPEEWEKTKK 80
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
Protein:
yrdC;BU494;hypothetical protein|543652..544179(comp)
Length: 175
N-terminus: OUT
Number of transmembrane
helices: 0
Transmembrane helices:
Total entropy of the
model: 17.0159
Entropy of the best
path: 17.0159
The best path:
seq MLHNDDVIAY
PTESMFGLGC DPNSKKAVKK LLNLKNRSIE KGLILVASDF
50
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq DQIKMYVNEN
ILSNKQKKKI FFHWPGPFTF LLPAKPNTPY WLTGKFNTVA
100
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq VRVSAHFEII
KLCNAFGQAV VSTSANISNM TPCFTSEEVF KCFGKDFPLL
150
pred OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO OOOOOOOOOO
seq NGKIGNEKNP
SKIINIINGK LIRYV 175
pred OOOOOOOOOO OOOOOOOOOO OOOOO
If you are going to use
these results in your work, please cite:
* G.E Tusnády and I. Simon (1998) /J. Mol. Biol./ *283*,
489-506.
* G.E Tusnády and I. Simon (2001) /Bioinformatics/ *17*,
849-850.